|
View web version
|
www.douglas.co.uk
|
|
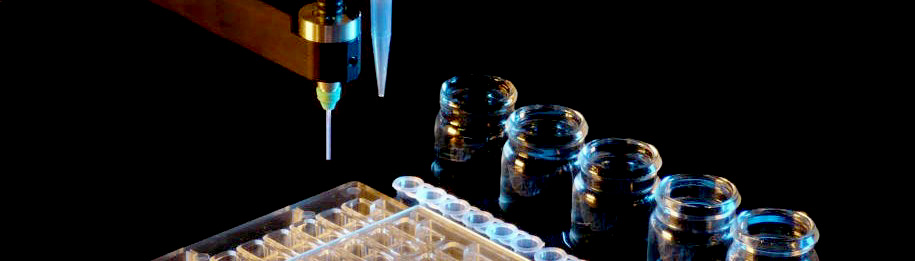
New plate for optimization now available from Douglas Instruments
|
|
The MRC Maxi optimization plate is a new design for macromolecular crystallization presented in a 48 well format.
The Oryx range of crystallization robots can easily set up 48 large (4+4 µL) sitting drops into this plate.
Just type the volumes that you would like into the experiment designer.
Manufactured by SwissCI AG, the plate offers an SBS footprint while providing 48 wells.
The MRC Maxi is intended for large drops and is compatible both with standard robotic systems as well as manual pipetting.
|
|
MRC Maxi - advantages in brief
Easy crystal retrieval
Raised wide wells make crystal mounting especially easy.
Easy viewing
Each well has a micro lens for perfect illumination. Micro numbering readable under the microscope for each well. (UVP) is UV transmissible.
Recommended volumes
Volumes validated for MRC Maxi are up to 10 μL of sample drop and 200 μL of the crystallization reagent.
Perfect for Optimization
The MRC Maxi plate is ideal for microlitre sized optimization experiments and is available both in polystyrene (PS) and UVP.
|

|
|
|
Using Oryx robots and our screening software WaspRun crystallizers can set up many types of experiment with the MRC Maxi plate including Microseeding,
Additive Optimization, Scaling Up, 4 Ingredient Gradients and Cross Matrix Designs.
Oryx 8 also has the benefit of X-Step optimization software. Users can dispense screen
solutions to the reservoir wells. X-Step allows users to quickly design gradient type optimizations or up to 7 dimensional
multivariate experiments.
|

|
|
|

|
|
|

|
|
|

|
|
|
Technical tip: Conversion of Vapor Diffusion experiments
to Microbatch-Under-Oil
|
|
A challenge for crystallographers wishing to use the microbatch-under-oil approach
is how to translate a known crystallization condition from microbatch to vapor diffusion, or vice versa.
Douglas Instruments recommends the following 3 steps for converting vapour diffusion drops to microbatch under oil:
1. Precipitant concentration is normally lower in the microbatch drop than in the vapor
diffusion reservoir. Set up microbatch trials covering a range of concentrations from
just below the concentration used in the reservoir, to half of this concentration.
2. Protein concentration is normally slightly lower than the stock solution used in
vapor diffusion (i.e. the protein concentration before mixing it with well solution).
Test a range from the concentration of the vapor diffusion protein stock solution to half this concentration.
3. In cases where crystallization takes place very rapidly, or where equilibration is slow
(e.g. using high concentrations of polymers such as PEG), crystallization may take place in vapor diffusion before the
diffusion equilibrium is complete. In such cases, the microbatch concentrations of protein and precipitant may be close
to the concentrations in the vapor diffusion drop immediately after setting up (before equilibration takes place).
for more on this see our website
|
|
Recent citations of Douglas Instruments products
|
|
The structure of a novel glucuronoyl esterase from Myceliophthora thermophila gives new insights into its role as a potential biocatalyst
M.-D. Charavgi, M. Dimarogona, E. Topakas, P. Christakopoulos and E. D. Chrysina
Acta Cryst. (2013). D69, 63-73
High resolution crystal structure of Sco5413, a widespread actinomycete MarR family transcriptional regulator of unknown function.
Holley TA, Stevenson CE, Bibb MJ, Lawson DM.
Proteins. 2013 Jan. 81(1):176-82
The Escherichia coli Lpt Transenvelope Protein Complex for Lipopolysaccharide Export Is Assembled via Conserved Structurally Homologous Domains
Riccardo Villa, Alessandra M. Martorana, Suguru Okuda, Louise J. Gourlay, Marco Nardini, Paola Sperandeo, Gianni Dehò, Martino Bolognesi,d, Daniel Kahne and Alessandra Polissi
J. Bacteriol. March 2013 vol. 195 no. 5 1100-1108
Crystallization of SHARPIN using an automated two-dimensional grid screen for optimization
B. Stieglitz, K. Rittinger and L. F. Haire
Acta Cryst. (2012). F68, 816-819
Structural and Functional Characterization of NikO, an Enolpyruvyl Transferase Essential in Nikkomycin Biosynthesis
Gustav Oberdorfer, Alexandra Binter, Cristian Ginj, Peter Macheroux,1 and Karl Gruber
The Journal of Biological Chemistry. (2012). 287, 31427-31436
`Seeding' with protease to optimize protein crystallization conditions in in situ proteolysis
J. Huang, Y. Gong, D. Huang, L. Haire, J. Liu and Y. Peng
Acta Cryst. (2012). F68, 606-609
Use of a repetitive seeding protocol to obtain diffraction-quality crystals of a putative human D-xylulokinase
R. D. Bunker, J. M. J. Dickson, T. T. Caradoc-Davies, K. M. Loomes and E. N. Baker
Acta Cryst. (2012). F68, 1259-1262
|
|
|
|
|
|
If you would like further information or have any questions about the topics in this newsletter, please contact me:
Tel: + (44) 1488 649090 US toll free: 1-877-225-2034
or Stefan@douglas.co.uk
With best regards,
Stefan Kolek
|
|
To request a service please contact Stefan@douglas.co.uk.
To request a demonstration please contact Hilary@douglas.co.uk
|
|
Douglas Instruments Ltd. All rights reserved.
Douglas Instruments Ltd
East Garston
Hungerford
Berkshire RG17 7HD
United Kingdom
Tel: + (44) 1488 649090
US toll free: 1-877-225-2034
|

|
|
|
|

|
|
Do you
have a comment or question about protein crystallization or robotics that
members of our bulletin board may be able to help you with?
Subscribe
to the Automatic Protein Crystallization Group
|
|
|
Visit this group
|
|
|
|
Douglas Instruments
|









