
|
|
See also: random Microseed Matrix Screening 'rMMS'IntroductionCrystallization is often divided into two steps: in the screening step, solutions that have previously given crystals are used to find new conditions where a protein crystallizes. Then, in the optimization step, the resulting crystals are improved by making small changes to the crystallization conditions identified in the screening step. Microseeding has been used routinely for optimization by a minority of crystallizers for many years (Bergfors, 2003). However this is painstaking work because you need to identify conditions where seeding is likely to work. A new approach falls somewhere between screening and optimization. It's very easy, and it often picks up entirely new conditions - conditions where crystals would not form in the absence of crystal seeds. The quality of the crystals found is generally better too. BackgroundIreton and Stoddard (2004) introduced a new approach to microseeding, which they dubbed “microseed matrix screening”. Crystal seeds were systematically added to diverse conditions as part of the optimization procedure. These conditions included ingredients that were not present in the original hits. For example, these authors identified crystallization conditions for the protein yCD that contained sodium acetate in a standard screening experiment. The resulting crystals, however, could not be used to solve the structure of the protein because the mosaicity was too high. However, by seeding into conditions where the sodium acetate in the initial hit has been replaced with calcium acetate, well-diffracting crystals were grown that allowed the structure to be solved. Surprisingly, crystals could not be obtained in the calcium conditions without microseeding. Microseeding was carried out by “streak seeding” using a cat’s whisker. D’Arcy et al. introduced two practical changes to
this approach, which together allowed automation: (1) seeding experiments were
carried out using ordinary commercial crystallization screens. (2) Microseeding
was carried out using solutions containing seeds prepared by crushing crystals
using the “seed-bead” kit from Hampton Research. Douglas Instruments wrote a
script that allowed vapor diffusion experiments with microseeding to be set up
automatically and with minimum manual intervention. These experiments were
identical to ordinary vapor diffusion experiments except that 0.1 ul of seed
solution was added to each drop. The seed solution was picked up from a
PCR tube on the robot stage. This "random microseed matrix screening" (rMMS)
approach increases the chance that crystals will grow in the metastable zone of
the crystallization phase diagram. InterfaceThe figure below shows the simple interface for designing rMMS experiments with Oryx8, Oryx4 or OryxNano. The user can choose the plate, the volume of reservoir solution to be added to the drop, the volume of protein in each drop, the volume of seeding solution etc. 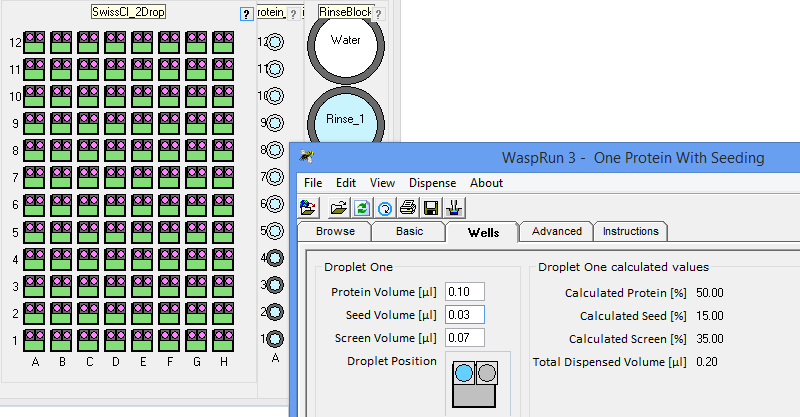
ResultsD’Arcy et al. (2007) reported that the average number of hits obtained for 5 target proteins increased by a factor of 7. Other users have reported similar improvements. For example Lesley Haire (NIMR, UK) obtained 6 hits in a screen, all poorly formed. After using rMMS, around 30 hits were obtained, including several well-formed crystals. 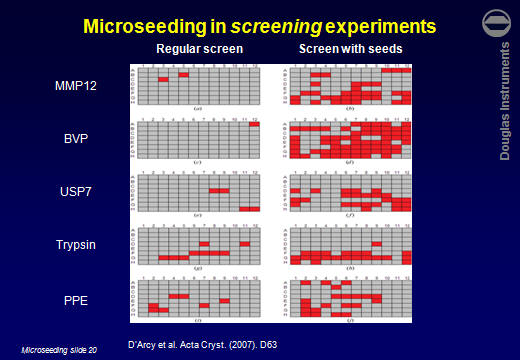 Jens-Christian Navaro Poulsen (University of Copenhagen) obtained 1 hit in 288 wells. After using rMMS, 10 hits were found in 96 wells. Then, using one of those crystals for a second-generation rMMS experiment, a further 10 hits were found in another 96 wells. Laura Cendron (University of Padova) used the same approach with a protein that crystallized easily, but tended to give small crystals. The best crystals obtained without microseeding are shown on the left. After microseeding, several good crystals were obtained (right).
TheoryFigure 1 shows a schematic phase diagram for a protein crystallization experiment. At high protein and high precipitant concentrations, precipitation occurs as shown. At low protein and low precipitant concentration, the solution remains clear. Near the bottom of the precipitation zone, there is an area where protein crystal nuclei form and grow (labeled "nucleation"). Below this is an area often called the metastable zone where crystals will grow, but no nucleation occurs. If you put a crystal (e.g. from the nucleation zone) into this area it will grow. However, if you set up conditions in the metastable zone without adding crystals or nuclei, no crystallization will take place. You can think of the wells of a typical screening experiment as arrows on the phase diagram. Water is removed from the drops, so these arrows point away from the origin (water is at the origin of this diagram). In a conventional screening experiment crystals are only seen if the arrows move through the nucleation zone, indicated by the blue and green arrows. The best results come if the condition moves a short distance into the nucleation zone (green arrow). Here only a few crystal nuclei are formed, so they are more likely to grow large. If the nucleation zone is small, crystallization may be a rare event because few arrows may land in the nucleation zone. By adding microseeds to the experiments, crystals can grow in experiments
that never leave the metastable zone, shown by the red arrows in Figure 1.
Note that the seed stock contains very little protein, i.e. it is on the bottom right of the phase diagram above. This means that the micro-seed crystals are unstable. Therefore it is very important to keep the seed stock on ice during the experiment and to freeze it as soon as possible afterwards. Note that in the case of yCD with calcium acetate, Ireton and Stoddard were unable to find conditions where spontaneous nucleation occurred - i.e. there was no nucleation zone. This may explain why such a great increase in the number of hits has been seen using the rMMS method, because some hits are found that could never have been picked up using conventional methods. Such cases may be ideal for growing crystals for data collection because the number of crystals can be controlled very exactly by controlling the number of nuclei that are added to the well. From these considerations it can be seen that the rMMS approach has three important advantages:
References
Microseed matrix screening for optimization in protein crystallization: what have we learned?
Protein crystallization with microseed matrix screening: application to human germline antibody Fabs
(Open-access)
Structure of arylamine N-acetyltransferase from Mycobacterium tuberculosis determined by cross-seeding with the homologous protein from M. marinum: triumph over adversity'
Random microseeding: a theoretical and practical exploration of seed stability and seeding techniques for successful protein crystallization
Promoting crystallization of antibody-antigen complexes via microseed matrix screening
(Open-access)
An automated microseed matrix-screening method for protein crystallization
Microseed matrix screening to improve crystals of yeast cytosine deaminase
Seeds to Crystals
Click here for instructions for making the seed stock. Any comments or questions? Click here Request an online presentation about Microseed Matrix Screening
|
|